Thoughts on How to Build the Virtual Cell with Artificial Intelligence
my comments various papers on how to build a virtual cell
Thoughts on How to Build the Virtual Cell with Artificial Intelligence: Priorities and Opportunities
… [J]ust as mathematics turned out to be the right description language for physics, AI may turn out to play a similar role for biology - Demis Hassabis, CEO of DeepMind
A very interesting position preprint on how to build a virtual cell has been published on arxiv: https://arxiv.org/abs/2409.11654. I’ve previously came across this idea of virtual cell from DeepMind CEO Demis Hassabis: https://www.youtube.com/watch?v=AuO0Y8Iwq0E. Since DeepMind built AlphaFold, a DNA sequence 🧬 to 3D protein protein structure model, I guessed that its research team would be trying out to build the virtual cell by synthethizing a population of proteins interacting with each other. In the end, a cell is technically a bag of proteins interacting with each other 🦠.
First impressions on the preprint
There are many novel ideas proposed throughout the preprint. One of the main approaches proposed in this position paper is to build a foundation model (let’s call it AI Virtual Cell) which can produce embeddings at multiple scales: molecular, cellular and tissue-level universal representations. Such foundation model would be trained with multimodal data spanning genomic information (sequence data), fluorence microscopy (imaging), single-cell RNA sequencing data (sequencing data), multiplex imaging (imaging), spatial transcriptomics (spatially-resolved sequencing data, or imaging + sequencing data), among others. See the figure below
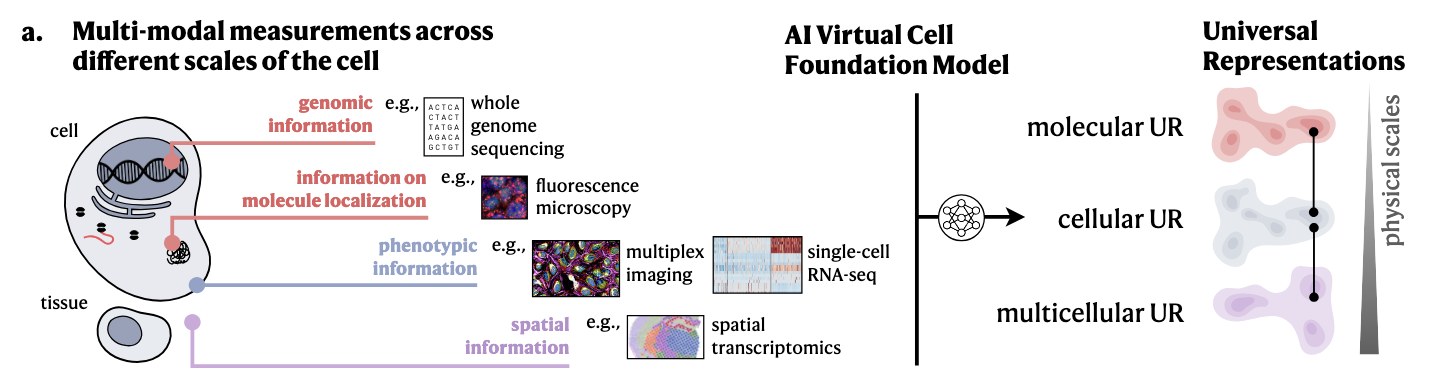
I understand it that such universal representations, which are essentially feature vectors $f \in \mathcal{R}$, can be used for some downstream tasks. The authors call the (neural network-based) models which use these feature vectors as “virtual instruments”, akin to digital tools🔬 to run in-silico experiments 🧫. If you have some temporal dataset that shows how genes change through mutations during metastasis, the idea is that you can use the AI Virtual Cell to obtain feature vectors of the genetic sequences, and train a network to predict the changes of genes across time.
While I understand that the need of feature vectors is ubiquituous for any machine/deep learning task, I disagree this is enough to build a virtual cell. On one hand, there are standard caveats to representation learning and building foundation models that we must be wary of. First, we have no control on what artifacts of the dataset a deep learning model is learning, and in biology it’s extremely difficult to distinguish noise versus actual biologically meaningful perturbations. The training of such foundation model would have far-reaching, biomedical implications, and as such extra-care must be taken for training this AI Virtual Cell embedder.
My other problem is that this position paper seems to mainly rely on statistical approaches to build this virtual cell and it’s constrained to building a model to produce representations, a form of bottom-up learning from biological raw data. It’s true that mechanistic modelling has limitations such as needing prior expertise/assumptions constrained to a simplistic setting such as modelling the growth of a population of cells assumming they want to replicate. However, I think mechanistic modelling, also known as a top-down modelling, will play a bigger role in building a virtual cell. Prof. Xavier Trepat offers a very good account on the complementarity of top-down and bottom-up processes to build a virtual cell. Consider the following thought experiment to note the importance of both approaches: top-down modelling allows us to explain high-level processes like traffic jams. While bottom-up processes are powerful since it allows us to thoroughly simulate and understand how a single car work, it at most help us know how a single car work, not necessarily how a population of cars lead to high-level problems. In biomedicine, if we want to further scientific discovery, mechanistic insight is indispensable for its interpretability and explanations to high-level phenomena. In that regard, deep learning and mechanistic modelling can complement each other to build a virtual cell.
Neuro-symbolic AI for building next-generation virtual cells
In one of the most interesting articles I’ve read recently on Building the next generation of virtual cells to understand cellular biology, I think a paradigm that can merge deep learning with mechanistic modelling⚙️ is neurosymbolic AI, in particular neurosymbolic programming.
Instead of focusing on training foundation model embedders, we can think of training a model that can output programs, where programs take as input raw biological data like a genetic sequence, and outputs a sequence of programs that can transforms the raw biological data. In the metastasis example above, the programs could explain to us how metastasis happens. See the depiction below:

Inverse reinforcement to model cell behavior
Another approach to building a virtual cell which I find very interesting and promising is by integrating inverse reinforcement learning (RL) as argued in this article: https://www.frontiersin.org/journals/systems-biology/articles/10.3389/fsysb.2024.1333760/full. One caveat in mechanistic modelling is its strong assumptions, for example, “cancer cells metastasize because they want to replicate”. This is disputable, what if the goal of cancer cells was different, and more nuanced than intially assummed. Inverse RL can alleviate this modelling assumption by not specifying some reward for a particular goal, but derive the reward function from observe behavior from cancer cells. The above paper proposes the following diagram, in which the first step on transforming the microscopy image to a cell state can make use of the above AI Virtual Cell embedder.
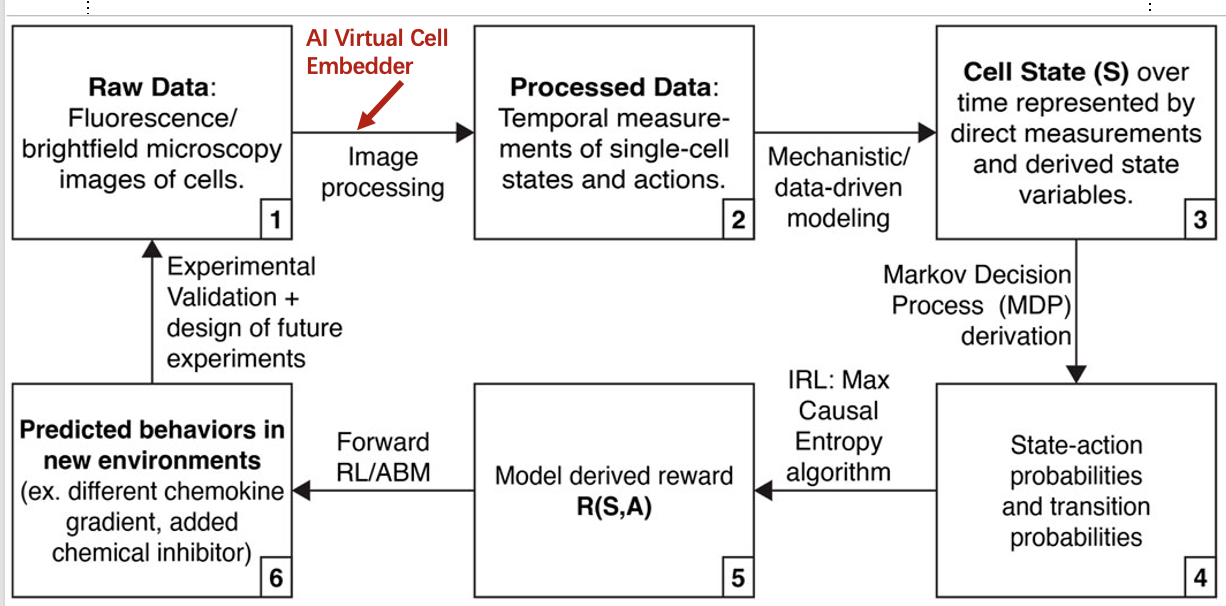
There are many approaches to building a virtual cell, and I think a combination of the above methods: deep learning, neurosymbolic AI and reinforcement learning, among others, will be needed together to build a virtual cell 🦠. How exciting times we’re living⚛. What do you think of the virtual cell?
A brief thought on serendipity 🧠🪐
Many methods in AI like neurosymbolic AI or reinforcement learning are not originally designed to solve biomedical tasks like single-cell data analysis, in a way such as a sequence alignment algorithm is tailored for identifying similarities in gene sequences or a deep network architecture is assembled to solve a medical imaging reconstruction task. At its core, research in AI is to understand how the brain works. Despite pursuing how human intelligence functions, this can nonetheless can yield unexpected insights into tackling problems in completely unrelated fields like network biology or cell science.
The above statement is supported by anecdotal evidence where scientific research translated into overarching technological applications for social good on completely unrelated fields. My confidence on such an idea can come from Prof. Geoffrey Hinton’s interview with CBS, among the endeavors of other researchers pursuing science whose work was converted into technologies benefiting humankind. Efforts include:
- Prof. Katalin Karikó, whose pursuit in understanding the central dogma of biology, namely how messenger-RNA translates into proteins, laid the foundation for the design of mRNA vaccines against the coronavirus during the COVID-19 pandemic.
- Prof. Yann LeCun, Prof. Yoshua Bengio, Prof. Jürgen Schmidhuber and Prof. Geoffrey Hinton himself, whose research into how the human brain works, such as how to simulate the visual cortex, how to reproduce human language, how can a machine achieve self-reference or how machines can learn like human beings, kickstarted the Third Revolution of Artificial Intelligence that can translate into a plethora of applications spanning over biomedicine
- Prof. Noam Chomsky’s work on a universal grammar underlying Human thought for understanding human language, which helped inspire work like the design of the FORTRAN programming language
- And many more examples, where who knows how research on intelligence can help with simulating a biological cell.
If the above anecdotes remind you of the book Why Greatness Cannot Be Planned by Prof. Kenneth O. Stanley and Joel Lehman, then there is another perspective from which to support the idea that basic science can fuel technological progress. We don’t know what are the stepping stones for achieving technological progress. Therefore, to achieve progress, we may have to be creative in the questions we ask and formulate novel ideas hoping and rely to some extent serendipity, that some of them will translate into groundbreaking innovations.