miRNA: General Blood Biomarkers
Research on a blood biomarker that can be used for the prognosis of diseases like cancer and Alzheimer, and how it relates to artificial intelligence with medicine.
Preamble: paradigm shift in the field of medicine due to artificial intelligence
It is not the strongest or the smartest who survive, but the one most adaptable to change
- Charles Darwin
As Prof. Darwin once shared with this world, those that live to witness the continuity of history are not necessarily the strongest or the smartest, but the one that can readily adapt to the changes of environment. As for this teaching, it may well not only be targeted at humans only, but also to technologies too.
Throughout the course of history, new technologies (be it in the form of innovative techniques, products or services) have constantly substituted their predecessors. For instance, thanks to the development of the steam engine, internal combustion engine, gas turbines and electric motors, humankind was able to leave behind the use of horse carriages as means of transport and instead employ airplanes, high speed rails, cars, steamships
Another story worth sharing goes as follows: in the early stages of the development of artificial intelligence, a rules-based approach was used, that is hand coding a set of specific task-oriented algorithms for the system to solve a problem, so for instance, natural language processing was a task performed through implementing a set of grammar rules. This approach was then gradually substituted by an upgrade in learning techniques for computers: machine learning (for the above case, instead using a statistical approach to analyze large corpora of texts to extract rules), where generic algorithms that could resemble human learning were preferred for machines to become robust when designed to solve certain task. This technique then was adopted through neural networks, from which a plethora of novel techniques and architectures stemmed: deep neural networks, convolution-based networks, recurrent-based variants, generative adversarial networks1, among others. These networks tend to require an enormous amount of training data to be able to extract relevant features and adjust the necessary parameters to perform small tasks such as image recognition, language translation or speech processing. Such procedure could be called “big data for small tasks”. Nonetheless, it can be further improved as it doesn’t resemble biological neural network’s ability to be able to exploit “small data for big tasks”, such as abstraction, higher dimension reasoning, creativity, among others. So the next step or the next upgrade to be pursued in the field of AI can be, as Prof. Song Chu would term it, to inquire into the “dark matter” regarding human cognition and how it can be applied to AI2. As an example of such approach, by late 2017, Prof. Geoffrey Hinton, who is regarded as the creator of deep learning3, proposed the concept of “Capsule Networks” as an alternative to Deep Learning that could perform better in image recognition or video processing with less data
The above anecdotes may differ in content, but an overall common denominator they share is the pursuit of a technological innovation because from such upgrade a plethora of techniques may bloom from that may revolutionize the way we live.
A field in which I want to pay particular attention is in medicine, specifically in personalized diagnosis and prognosis, as well as therapy of diverse diseases and how AI can merge with this field in order to improve the accuracy rate of diagnosis as well as develop targeted therapy. There are already notable achievements in this hybrid field, such as:
- Convolutional neural networks trained on medical imaging that can outperform radiologists, ophthalmologists, dermatologists
- Neural networks trained on electrocardiogram readings to predict the possible onset of a heart attack; or trained on genomic datasets
- Natural language processors to act as a virtual psychotherapist or automate the reading and summarization of research paper
- Virtual records to constantly track the gut microbiome based on food intake to adequately balance the amount of nutrients to take, among others.
Of course, these advancements come along with their respective drawbacks, such as research in the lab setting and applications in the real world are not balanced yet, and some fields deserve further understanding as well as employ larger and more diverse datasets
However, most research regarding diagnosis as well as therapies of diseases tend to focus on the phenotype related to a disease, that is, what are the physical, observable symptoms of a patient, i.e., form of a skin rash, certain tumor’s size, symptoms such as cough, chronic distress, inflammation, pain, among others. So, for example, medical imaging seems to rely heavily on recognition of features in a lung scan or features of a skin rash to make an accurate diagnosis.
An innovation in this field, therefore, concerns the exploitation of information carried by the genotype related to a disease and, indeed, currently, we are seeing great advances and focus on the research of the genome related to a disease, diving into the realms of genomics and epigenetics to study disease and manufacture possible therapies. This area of study spans from genetic sequencing5 to identification of genetic biomarkers of a concerning disease, as well as genetic engineering for the development of targeted therapy for that particular disease.
Given the above introduction, I hope I’ve convinced you of the importance of identifying what are the current technological upgrades in the diverse fields of study such as AI. We can ponder on ourselves: what technologies will keep up with the fast pace of development and replace their predecessor, akin to how only adaptable species nowadays remain? How can we, as stargazers of the future, protagonists of the incoming generation do and act in order get hold of those technologies and catch up with these advances and development, to keep up with the current flow of history, a swirling torrent?
In medicine, I believe that it’s diving into the realms of genomics for early diagnosis and targeted therapy of a disease, and the biomarker presented here in the articles to follow is called microRNA and how it might play a crucial role in the development for an upgrade in this field.
For a brief sneak peek of what might the ripening of the study of how microRNA can impact the future of medicine, here is a hypothetical dialogue:
Doctor (calls): Hey Edmund, from the blood sample you provided yesterday and processed through the mirOculus (depiction) at your home, I can see that the results of your microRNA profiling, particularly an upregulation of microRNA-122 that suggests you have Hepatitis C.

Edmund (exaspirating): What? Doc I’m so worried.
Doctor: Don’t be worried since we caught it early. You can come to the hospital and we can perform a subcutaneous injection consisting of oligonucleotides that can inhibit the upregulation of that microRNA. This should disrupt the progression of the disease and provide long-lasting antiviral activity. Don’t forget to constantly monitor the microRNA pattern of you and your family.
Edmund (calmed down): Okay… thanks Doc, that was… pretty straightforward…
For those gazing at the future, I present microRNAs, a molecule which might become the next blockbuster in the era of the merging of medicine and AI by playing crucial roles in the early diagnosis as well as targeted therapy for diverse diseases. Of course, I have to clarify that microRNA targeting is just one of a myriad of innovative technologies to keep an eye one, analogous to only one boat from a crew that can carry us, young passengers, through the turbulent tides of technological revolutions that can only move forward.
Interlude: A brief introduction to what microRNA is
To materialize the dialogue above into reality, it is relevant to go through a fundamental overview of what miRNA is. Afterwards in the next section, we will dive deep into its applications in disease diagnosis and therapies. Because this interlude serves simply as an introduction to this nucleic acid, it’s written for those who are new to micro-biology and simply want to explore. As the reading progresses, the difficulty increments and a more technical vocabulary as well as more complex topics will be dealt with (topics that I do not thoroughly understand either).
microRNA regulates gene expression
Have you ever wondered why doesn’t your hair grow from head to toe in just one night? How about why doesn’t your nail grow 1–2 meters long? In the end, we just need your cells to express a large amount of proteins (such as keratin) and our nails would grow touching the floor in a short period of time. Speaking about cellular expression, we’ve been told that almost all 3.6 billion somatic cells in our body (excluding sexual cells residing in our reproductive organs and few other exceptions) are mostly similar: they have similar genetic code, mostly the same DNA sequence and coiled into chromosomes residing inside the respective nucleus6.
Indeed, they’re all the same… except they just aren’t: cells circulating in our blood vessels, those that make up the blood vessels, those constituting lung tissue or brain tissue, are clearly different from one another not only in terms of structure, but also in function, e.g neurons transmit electric signals, alveolar cells are in charge of exchanging oxygen and carbon dioxide between lung and blood, white blood cells carry diverse functions from producing antibodies (helper B cells), to inhibiting pathological activity through killing infected host cells(killer T cells), to ingesting pathogens and producing antigens that start an immune response, among others. Nonetheless, it remains true that they all carry the same genetic code. The difference is that each type of cell’s DNA sequence “differentially” expresses its respective genes which translates into different proteins, and these proteins either make up the distinctive structure that distinguishes cells among one another (only neurons have a myelin sheath for example) and represents their distinctive functions (only helper B cells produce antibodies that bind to pathogens’ receptors to inhibit their activities). The “decision” on what cells will constitute brain tissue, which ones the liver tissue, or which ones bone tissue can be traced back at your birth, where there existed a single mother cell (stem cell) that through differential expression of its genes ramified into the different cells, each carrying its distinctive structure performing their unique functions7.
So now we sorted why cells are different (their genes express different proteins) but still haven’t answered what mechanisms control which portion and how much of it is expressed. That is, the first questions raised above that concerned why hair or nails don’t grow exponentially all of a sudden remain unanswered. Simply speaking, this is because there is a mechanism that regulates gene expression, and this is a very complicated and heterogeneous mechanism. Gene expression, also referred to as the central dogma of biology8, is the process in which a sequence of nucleotides in the DNA is transcribed into mRNA and then translated into amino acids that form a protein. In summary, it happens as shown in (Figure 1):
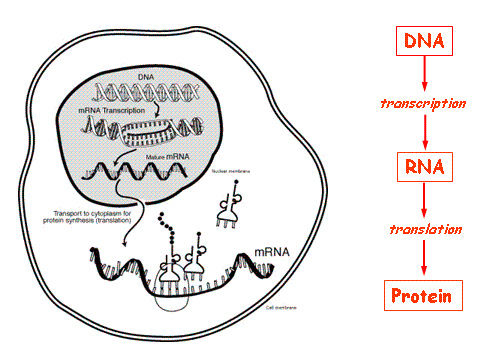
In the regulation of gene expression, micro-RNA (miRNA) plays an integral role. MiRNA is also a byproduct of DNA transcription (as explained later in this section), however it doesn’t undergo translation like mRNA; instead it inhibits the completion of the central dogma by pairing and inhibiting mRNA so that it’s unable to bind with tRNA anymore or degrades in the cytoplasm. Because it’s not translated, miRNA is also called noncoding RNA (nc-RNA).
So, when there is a balanced concentration of miRNA in the cell’s cytoplasm (not necessarily in the nucleus), mRNA translation can be regulated; conversely, if it isn’t, then this deregulation of concentration might decay into several diseases such as cancer, as dysregulation might mean the lack of proteins essential to maintain cell survival (such as pathways that induce apoptosis to prevent tumorigenesis) or the excess of proteins harmful for the cell, such as those that induce cell proliferation or prevent apoptosis.
So back to the original (strange) question proposed as a starting point to introduce miRNAs, the possible reason that may explain why our hairs or nails don’t grow exponentially, or as in general why our cells don’t over-produce proteins or undergo excessive replication (unless it’s part of a disease like cancer), it’s partly due to the balanced concentration of miRNAs that are constantly regulating gene expression.
With the above introduction, it is pertinent to talk about the biogenesis of miRNA. This is because it will serve as a premise for understanding how biomedical researchers design drugs used to target or mimic miRNAs as a viable treatment for diverse diseases. Biogenesis concerns pathways that lead to the formation of mature miRNA, and drugs are designed to inhibit such pathways for a downregulation of miRNA or imitate the structure of miRNA for upregulation of it.
Biogenesis of miRNA
MiRNAs are small ncRNAS that are 18-25 nucleotides long
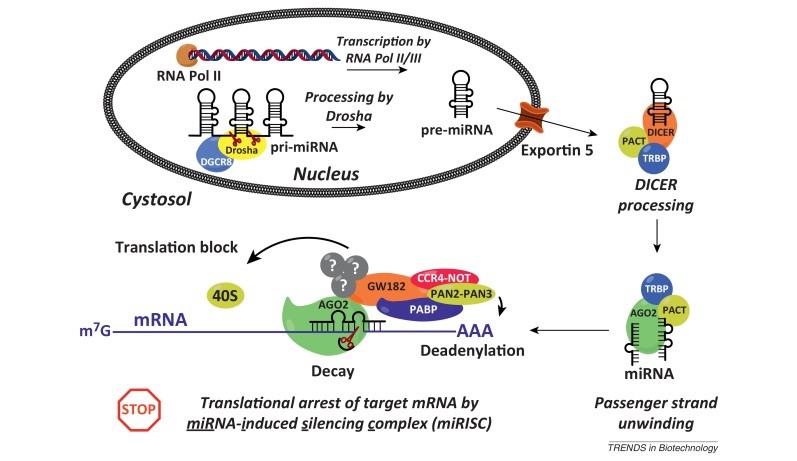
As stated above, miRNAs are also byproducts of transcription (however they apparently lack open reading frames, so they don’t get translated). Such transcription starts with RNA Polymerase III transcribing from the DNA primary miRNAs (pri-miRNA) that have hairpin structures. Afterwards, it is processed by a protein called DROSHA forming precursor miRNA (pre-miRNA), which are 70-100 nucleotides long. A protein called Exportin 5 recognizes this pre-miRNA and mediates its translocation to the cytoplasm
The description above corresponds to the canonical pathway (Figure 3a is the same as Figure 2), however, researchers have also found alternative pathways as displayed below.
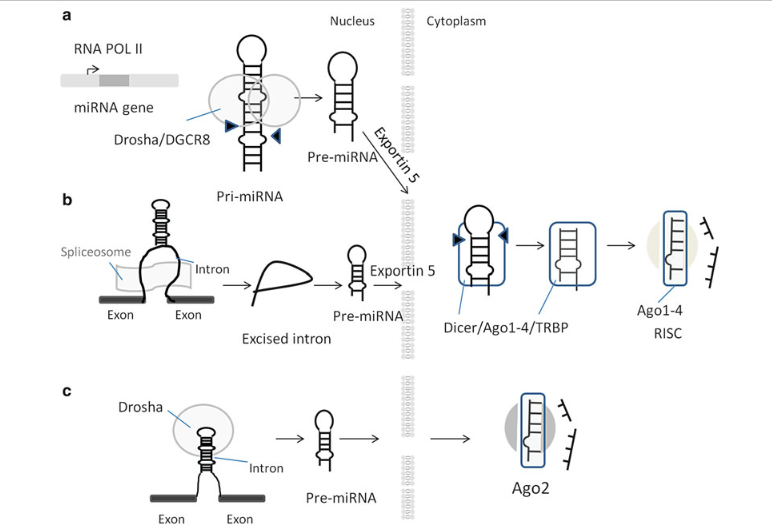
In Figure 3b, intronic microRNAs, also called mirtrons, are processed by another protein in the nucleus called spliceosome, an alternative to Drosha. These processed mirtrons, pre-miRNA, follows afterwards a similar path as above, that is, recognition by Exportin-5, and then processed into the RISC complex. Figure 3c describes simtrons, which in contrast to mirtrons, may require Drosha but not DICER protein complex processing
It is worth noting that previously it was thought that the unzipped 3p strand would have degraded in the cytoplasm, so its up or down-regulation was irrelevant for the study of the progression of diseases like cancer. Nonetheless, due to recent studies, it’s suggested that the 3’ complementary chain, or 3p strand, doesn’t necessarily degrade, but also plays a crucial role together with its complementary 5p strand in regulating gene expression. For example, the 5p and 3p strands of miR-582 and miR-28 are downregulated in bladder and colorectal cancer, respectively. However, experimental forced expression of miR-582-5p and miR-582-3p to achieve its upregulation inhibited bladder cancer cell proliferation and tumor growth, whereas forced expression of miR-28-5p, but not miR28-3p, reduced colorectal cancer cell proliferation
Despite given the above, it ought to be clarified that the study of these small molecules are still a developing field, so, for instance, the precise mechanisms involved in the miRNA transcription is not known
Regardless of such uncertainty, with what we currently know and will find out about miRNAs can already be used for diving deep into how it can act as a biomarker for the diagnosis (or maybe prognosis) of diverse diseases and afterwards for targeted therapy of such.
Dialogue: Voyage for a better health
Far away on the red sea, a big ship with a flag displaying “AGO–2” carrying brave Argonauts is seemingly wandering aimlessly.
Argonaut #1: Captain, when are we going to see some action? It’s been too long since we’ve performed a heroic deed in the faraway Red Sea.
Captain of AGO–2: no rush my brothers! Sooner we’ll come across a defective ship with a defective crew to stop them. Look, I can now see a ship from afar. Give me my binoculars, I need to see whether they’re acting normally or are they problematic.
The Captain stands from tip of the AGO – 2 and through the binoculars. The Captain sees that the crew members of that ship seem to be working as normal.
In the other ship:
Ship’s Boss: Alright, keep replacing parts of the ship and polish what’s dirty, so we can sail further into the Red Sea! Remember, we have to deliver the cargo by the end of today!
Crew members (shouting monotonously): Roger!
Crew members A, U, G (shouting): Boss, we’ll start changing the mast, polish the anchor, clean the deck, and lift new sails!
Ship’s Boss: That’s the spirit! Let our ship navigate further and more stable.
(After working for a while by replacing the defective parts of ship and performing other functions)
Crew members U, G, A (shouting): Alright, stop! That’s it, any more polishing or adding new parts is going to do more harm than good. Let’s just keep sailing further. Well done for today.
Back to AGO-2:
Captain of AGO–2: Hmm. That one seems to be working properly
Argonaut #2 (suddenly rushing): Cap’n! We’ve just spotted a ship whose crew members are working ferociously. They’ve already broken the mast and added new ones several times, polished the deck till it broke apart, built so many anchors and released them that the ship is now stuck. They’re crazy! Furthermore, they’ve added so many sails yet tore apart many too. Now their members are fighting each other and nobody seems to stop.
Captain of AGO–2: Oh my gracious! What’s gotten into their mind! From where do they derive such madness?
Argonaut #3 (looking from the deck): who knows? Such kind of reckless behavior seems to be probabilistic and happens quiet often.
In the maddened ship:
Corrupted member #1: I have to built anchors! Built more anchors! More anchors!
Corrupted member #2: add more sails! Add more sails! There is just not enough sails!
Mad captain: Stop adding more things to the ship! At this pace it’s going to sink!
Corrupted members #1 & #2: we can’t!
Mad captain: then jump off the ship!
Corrupted members #1 & #2: we can’t!
Mad captain: Bah! Humbug! Where is member #53 when you most need it?
Corrupted members #1 & #2 (building more things which add more weight to the sinking ship): we don’t know!
Back to the AGO-2 ship:
Captain of AGO–2: Drive AGO – 2 to the tail of that ship and let our members in. We have to stop this madness as soon as possible otherwise this ship is going to sink to the Red Sea and contaminate it.
Argonauts: Roger!
The tip of AGO–2 hits the tail of the ship serving as an anchor to keep both ships joint together and allow the Argonauts jump to the corrupted ship. The Argonauts jump the ship and start mediating the reckless behaviors of the corrupted members. Details are omitted.
After a while:
Captain of AGO–2 (looking from a binoculars from the deck of AGO–2): It seems that the ship is almost back to normal. The corrupted members’ behaviors have been regulated.
Argonaut #1: how did they do it?
Captain of AGO – 2: Well, can’t you see that “details are omitted”? Plus, I couldn’t even see clearly through my binoculars, since they were predesigned to be able to track the actions of only Argonaut #1301, who was in charge of convincing some corrupted members to commit suicide, and Argonaut #126, who seemed to have stopped such reckless behavior from spreading to other crew members. As for the specific, very specific details of how they did it, I can’t say for certain, and I hardly will, nonetheless, knowing that my fellow Argonauts can prevent the spread of such catastrophe is enough for me to ask of you to recruit more Argonauts like them.
Afterthoughts
Like before, I’d love to share some of my thoughts regarding the crafting of such section as well as the dialogue annexed to it. To begin, I do clarify that this was just an Interlude, meaning that it was just a “side story” to introduce you the concept of miRNAs and how they are created, in other words, their biogenesis, as this would serve as a basis for understanding how it can act as a biomarker for diverse diseases ranging from cancer to neurological diseases like Alzheimer. However, I didn’t, and won’t dive deeper into the specific details surrounding miRNA, like their structure, how “precisely” they work, partly because I’d like you to read it yourself from papers available online; the main source I consult is PubMED from NCBI. Another source, which I’ve mentioned in the bibliography is called “miRNomics: MicroRNA Biology and Computational Analysis” by Malik Yousef and Jens Allmer, which you can search online for a thorough explanation of miRNAs. As for my produced content, I’d instead focus on introducing to the reader what are the possible prospects to look up to in this emerging and amazing field in a way that’s understandable and creative.
Now, proceeding to the Dialogue: Voyage for a better health, AGO–2 is basically referring to the AGO-2 protein complex that carries the guide strand of miRNA, represented by Argonauts. They are sailing on the Red Sea which is analogous to miRNAs flowing in our blood. The first ship the Captain encountered was supposed to represent the moderated translation of mRNA, resembling the modulated growth of our hair, nails, or, simply speaking, not developing cancer, as shown by how crew members worked properly and didn’t proliferate the parts of the ship like the mast, sails or anchors. A little detail that you may have noticed is how crew members A, U, G signaled the start of the crew’s activity, which was an analogy to how the start codon signaled the beginning of mRNA transcription. Similarly, crew members U, G, A represented one of the 3 stop codons that ended mRNA transcription. AGO–2’s crew probably didn’t intervene as they seemed to work ordinarily. Conversely, the next ship was an example of a cancer. Its crew members started acting recklessly, building at an un-necessarily large scale parts of the ship that were doing more harm than good, which you can view it as tumorigenesis or angiogenesis. The captain of this corrupted ship asked why crew member #53 wasn’t here to stop the progression of this cancer, which resembles the lack of p53’s activity during tumorigenesis, where p53 is a tumor-suppressor protein that induces apoptosis in cancerous cells. It’s of course of great interest to understand why do crew members go reckless, but I answered it through Argonaut #3’s comment, which is that such occurrence, given that’s caused by genetic mutations, is probabilistic by nature. What happens afterwards till the end is analogous to what was explained, AGO – 2 anchoring itself to the corrupted ship resembles miRNA binding to the 3’ UTR of the target mRNA and, in this case, inhibiting its activity. The two Argonauts correspond respectively to miR–1301, which promotes apoptosis, and miR-126, which inhibits metastasis
The last comment by the Captain restates an idea shared previously, which is that we don’t know precisely how miRNA works despite they’re playing an essential role in regulating gene expression which correlates with cancer progression, so further studies are required; in the end, science is the art of reducing uncertainty; the use of a ship sailing to the Red Sea to convey the functioning of miRNAs is to transmit the underlining idea that science is a never-ending voyage of discovery… Nonetheless, with the current knowledge we already have, and researches being carried out, we already have at our disposal useful knowledge that can be put to practice in order to diagnose and treat diseases that involve miRNA deregulation.
So, for the next section we’ll focus on how miRNA can act as biomarker for diverse diseases and some thoughts about how artificial intelligence can play a relevant role for this field’s development.
DeepMiRNA: miRNAs as biomarkers for early diagnosis of diseases
“The best cure for any disease is its prevention”. A more down to earth paraphrasing can be the “best cure for a disease such as cancer and its diverse presentations like leukemia, thyroid carcinoma or other disorders like neurological ones such as Alzheimer, is their early diagnosis and subsequent targeted/personalized therapy. For the former, current approaches to achieve an early diagnosis for the diseases above mentioned are not as effective as expectations would demand. They mostly focus on the phenotype expressed by the disease, such as lung tumor size (lung cancer), skin rashes (skin cancer), identify the presence of protein complexes like beta - amyloid, that come with its disadvantages such as the difficulty to at a premature or early stage identify a primary cancer (which can be as low as 100000 cells undergoing mutations). Current approaches that involve artificial neural networks trained at image recognition in order to identify at an early stage features of diseases like cancer from radiographies has shown promising results by outperforming real ophthalmologists, radiologists and pulmonologists; of course, these innovative approaches hasn’t yet to meet a large scale real-world applications, and in practice they meet their limitations such as the data gathered such as electronic health records being stored in different formats so it can’t run on diverse software or not having a diverse enough dataset that’s able to capture health tendencies among distinct ethnicities
Given the described issues, the need to find novel, upgraded ways to deliver medicine is thus necessitated; in this case, it’s reinstated the an idea shared previously, one can now also focus on the genotype related to a disease.
Around 20 years since the creation of The Human Genome Project, the study of genomes has attracted researchers thanks to the seemingly causal relationship shared between pathogenesis and genetic makeup. Furthermore, as more understanding is built upon from the Human Genome, attention is shifted from protein coding sections to the nonprotein-coding RNAs, a field which have vastly expanded with ongoing research studies and now include transcripts such as Piwi-interacting RNA (piRNAs), long ncRNAs (lncRNAs) and microRNAs (miRNAs), the last one arguably are one of the most predominantly represented ncRNA groups in clinical research
The field of study of miRNA, which has acquired for itself the name of “miRNomics”
In contrast to diagnostic methods that may be invasive like biopsy or require taking samples that are not easily accessible like cerebrospinal fluid, miRNAs appear to be easily accessible given that they may circulate in extracellular vesicles called exosomes of the blood
Even diseases like Alzheimer seems to be able to be prognosticated through a miRNA signature detection despite being a neurodegenerative disorder which current diagnostic methods involve extraction of cerebrospinal fluid, for instance. This is because miRNAs are able to cross the blood-brain barrier, therefore can potentially be quantitated in routine blood, serum, or plasma tests as measures for, not only Alzheimer, but other neurodegenerative and neurodevelopmental impairments
As an additional point, it’s worth reinstating that the potential for miRNA-based diagnosis is better exploited when prognosticating a disease, as an early detection increases a patient’s survival. As a matter of fact, regarding Alzheimer, it seems from
miRNAs as potential biomarkers
The current literature mostly emanates an optimistic atmosphere regarding the measure of relative miRNA’s concentrations to prognose a disease. This is because several miRNAs have been discovered to be linked to the capability of regulating either cell proliferation, apoptosis, differentiation as well as diverse molecular pathways. Deregulation of these miRNAs (whether it’s upregulation or down-regulation) can lead to the development of a disease: examine for instance the handmade table containing the abstract of different papers that has identified a miRNA signature related to a disease:
| Source | Extract |
|---|---|
| Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2 | “Impressively, we found hsa-miR-4661-3p was predicted to target the S gene of SARS-CoV-2, and a virus-encoded miRNA MR147-3p could enhance the expression of TMPRSS2 with the function of strengthening SARS-CoV-2 infection in the gut” |
| Role of microRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease | “… miR-218-5p was significantly down-regulated in patients with COPD, compared to normal subjects” |
| MicroRNAs as Biomarkers in Thyroid Carcinoma | “Circulating levels of miRNA-146b5p, miRNA-221-3p, and miRNA-222-3p in PTC (papillary thyroid cancer) patients have been found to be higher than those in healthy controls…” |
| Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease | “Significant upregulation of miR-92a-3p, miR-181c-5p and miR-210-3p was found in the plasma of both MCI and AD subjects…” |
| Identifying a miRNA signature for predicting the stage of breast cancer | “… four miRNAs, hsa-miR-503, hsa-miR-1307, hsa-miR-212 and hsa-miR-592, were significantly associated with the prognosis of patients with breast cancer.” |
Through diverse methodologies that combine experimental, bioinformatics or statistical analytical approaches, researchers are constantly identifying useful causal relationships between RNAs and the phenotype of a disease. However, because of the ever-increasing amount of research output, it’s becoming impossible for a human being to keep up with this large tide of discoveries; in the end, there are about 2000 miRNAs identified so far, and it is believed that they collectively regulate one third of the genes in the genome
Despite such a dynamic, ever-evolving process of assimilation of discovered miRNAs, it appears that the fundamental task of identifying which miRNAs and how are they deregulated so that it causes the phenotypes that constitute the early development of a disease, remains unchanged. Due to this, this is where artificial intelligence can serve as a useful tool to aid reasearchers in extracting from the massively available papers relevant data about:
- How a miRNA’s up or down-regulation is related to the onset of a disease
- How that miRNA interacts with a biological pathway
- How that miRNA interacts with genes or other molecules such as another miRNA
- Among others
A viable approach is to train natural language processors to produce from inputs made up of research papers knowledge graphs that display the data about miRNAs laid out above. Such an idea has been employed by startup company miROculus (previously mentioned, which focuses on detecting circulating miRNAs’ concentrations to achieve an early diagnosis of cancer) is displayed in Figure 4:
According to a case study from Neo4j (a graph database), the miROculus team gathered more than a billion articles in Hadoop and used a NLP to extract specific sentences with keywords for gene, disease and microRNA and subsequently infer the relationship between keywords; the team developed an unsupervised machine learning model to classify relationships, which are then stored in Neo4j9. It’s also worth noting that it’s open-access tool for the science community, which opens the opportunity for research to support or refute any knowledge that’s displayed. Nonetheless, although it’s claimed that this interactive site is updated monthly
Similar approaches that compile miRNA research results from papers have been done, such as online databases (please visit http://www.mirbase.org/ or http://www.cancerindex.org/geneweb/index.htm ).
Accessing miRNAs biomarkers
Of course, if we inquire deeply regarding the potential of miRNA as biomarkers, apart from sorting out the causal relationships, another concerning issue is the methodology employed to access and isolate them for clinical analysis, which methods may vary in precision or ways to perform it completely. For instance, Table 2 shows some methods used to isolate and analyze miRNAs from a biospecimen like blood plasma or serum:
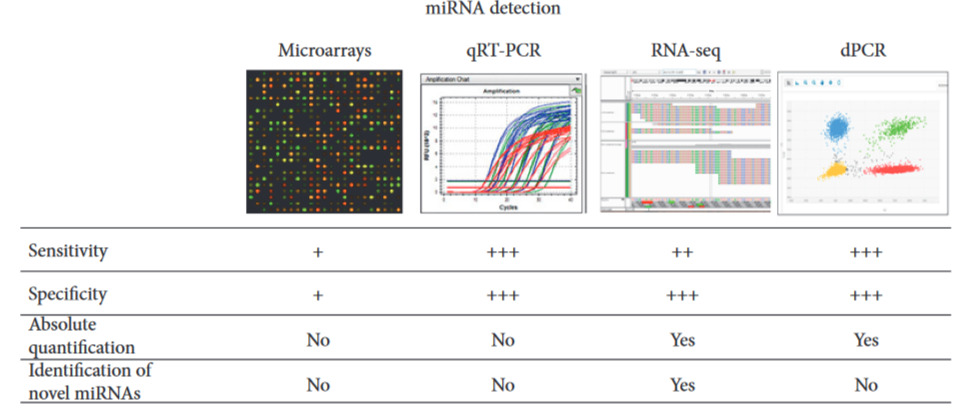
However, although the above methods are able to complete the task of detecting and measuring miRNAs, they come with perhaps essential limitations: they require bulky and expensive equipment, intensive sample preparation, or long turnover times, which limit most of them to well-equipped laboratories
Discussing about the simplicity of a method to detect and measure miRNA’s concentration is relevant as it influences whether it’ll be available to the general population or reserved to medical professionals or researchers. Issues to be taken into account are:
-
Cost of reagents and tool: ideally, to keep them as minimum as possible if the goal is to “democratize medicine”, borrowing the words from miROculus co-founder Jorge Soto.
-
Difficulty of methodology to process the (blood) sample and subsequent analysis: the ideal would be to design a method with limited sample preparation that a nonexpert could execute
, which would allow the capability to constantly monitor circulating miRNAs like how one would regularly track blood glucose and blood pressure from a glucose-meter and sphygmomanometer used at home. As such, the time taken to get the results of such measurements would also ideally be limited to within hours.
CRISPR-CAS System and miRNA diagnostics: search for a universal diagnostic method
CRISPR (short for Clustered Regularly Interspaced Short Palindromic Repeats) – CAS system was initially discovered in bacteria and archaea, as an acquired immune system against viruses and phages through CRISPR RNA (crRNA)-based DNA recognition and Cas nucleases-mediated DNA cleavage
However, recent developed techniques has also spanned to the field of nucleic acid detection. Such advancements have promoted next-generation diagnostics employing CRISPR-CAS systems such as Cas 12, 13, 14 by making use of their nonspecific degradation of non-target molecules (trans cleavage) after specific recognition of the desired nucleic acid
Regarding miRNA detection, there also seems to be great potential yet to be discovered from employing the CRISPR-CAS 13a system. Unlike qRT – PCR, a Cas 13a based detecting platform doesn’t need to amplify the target molecule, as well as it’s relatively more low cost, easily scalable (doesn’t require much time nor technical steps) and it’s highly specific, meaning that it can discriminate highly homologous miRNAs from the same family. Such advantageous qualities are shown, for instance, from the research from the South China Normal University as well as from University of Freiburg.
For the former,
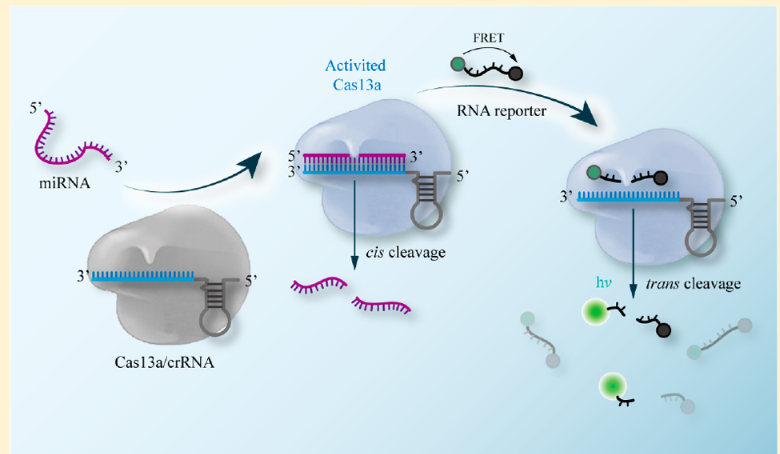
Reporter RNAs are mixed with the sample with/out the target miRNA. If the Cas13a system’s programmed crRNA detects the desired miRNA from base complementariety, Cas13a’s cleavage mechanism is triggered which indiscriminately cleaves RNA nonspecifically, including the reporters, releasing fluorescence signals. Fluorescence signal, hence, will be proportional to the concentration of the target miRNA.
For the latter,
All in all, the CRISPR-Cas system, whether it’s further polishing techniques using the Cas13a nuclease or another enzyme, holds a lot of promises for developing a detection platform to measure miRNA’s concentration at the point of care, which would made possible the early detection of a disease given that it’d compensate for the disadvantages of the previously mentioned methods such as microarrays, RNA-seq or qRT – PCR, which are restricted to be used in technical settings for research, lifting a barrier towards a wide-scale usage for health monitoring.
Causes of miRNA dysregulation
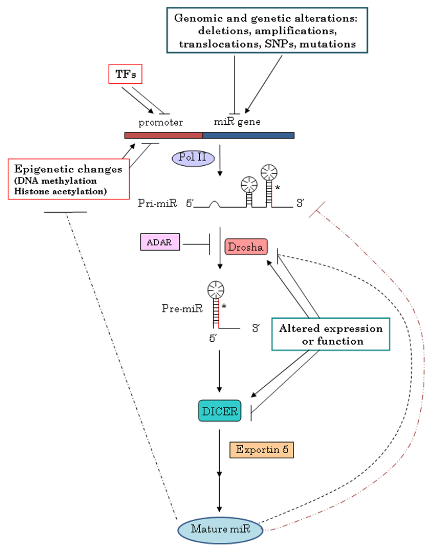
After sharing the above knowledge regarding miRNA deregulation and pathogenesis, interest might arise about why miRNA becomes dysregulated. To explain the identified causes of miRNA dysregulation, it’s first worth noting that miRNAs do not only regulate gene expression, but they themselves are also controlled by regulatory mechanisms, which may get altered during tumorigenesis, leading to either overexpression of oncomiRNAs or down-regulation of tumor suppressor miRNAs. Figure 6 summarizes the mentioned causes of miRNA dysregulation. As noted by
-
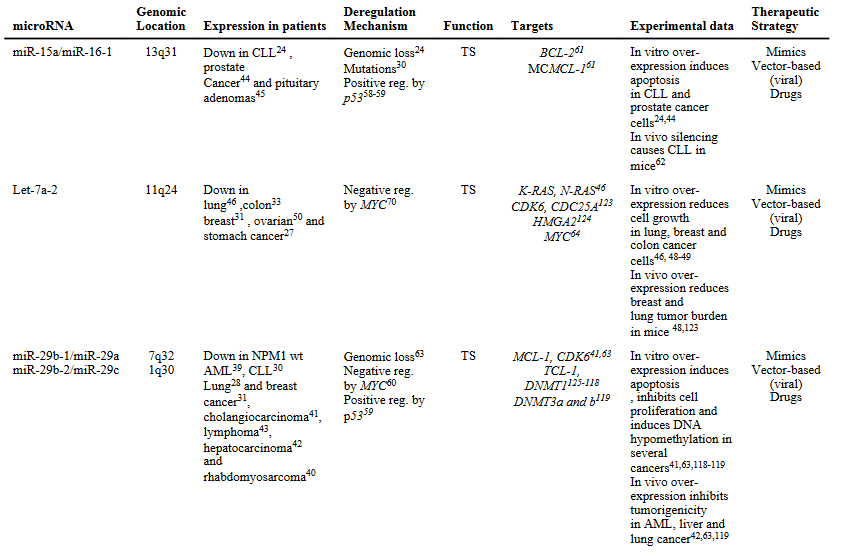
Mutations that involve genes that transcribe into miRNAs can affect the expression of such miRNA. For example, in Chronic Lymphocytic Leukemia, mutations lead to the reduced expression of miR-15a and miR-16-1. Another example of an oncogenes affecting miRNA expression is represented by the oncoprotein MYC which is able to both induce oncogenic microRNAs,as the miR-17-92 cluster, and negatively regulate transcription of tumor suppressor miRNAs, such as let-7 and miR-29 family members
-
Altered activity of transcription factors can also lead to miRNA’s abnormal concentrations. For example, an interesting regulatory loop has been demonstrated between ZEB1 transcription factor and miR-200 family: Epithelial-Mesenchymal Transitions (EMT) inducers ZEB1 and ZEB2 (both transcription factors) are direct target of miR-200 family members, and vice versa, ZEB1 has been shown to directly repress miR-200c and miR-141transcription.
-
Altered miRNA’s concentration can also be due to epigenetic changes, such as altered DNA methylation, with DNA hypomethylation possibly leading to upmodulation of oncogenic miRNAs, while DNA hypermethylation leading to the silencing of miRNAs.
-
Defects during miRNA biogenesis, such as for instance altered DROSHA or DICER activity. For example, it seems that the silencing of either DROSHA or DICER promotes tumorigenesis in vivo.
-
miRNA’s expression level can also be affected by other miRNAs, thus creating a complex level of reciprocal interaction and regulation; for example, in a mouse model by
, miR-709 seem to be able to bind to pri-miR-15a/16-1 preventing its processing into pre-miR-15a/16-1 and thus leading to a suppression of miR-15a/16-1 maturation.
Challenges of miRNA diagnostics
After having discussed about miRNAs being potential biomarkers and detection platforms used to measure their concentration levels, it is also worth noting what are the challenges so far seen in this field of study. To begin, despite the high amount of research published pinpointing what miRNAs cause certain disease, there are cases where proposed miRNA biomarkers differ for different research groups even though the same disease is studied; as an example please examine
Another challenge concerns the extraction of miRNA from a blood sample. As said before, miRNA from a blood sample may originate from extracellular vesicles called exosomes. Yet, there are investigations that probe whether enrichment of exosomes from biological samples are better than crude cell-free preparations for reliable biomarker measurements; and some reports suggest it may depend on the specific human disease. Vesicle purification can lower the RNA yield and integrity. Furthermore, some diseases may also alter exosome release and clearance, or administration of therapeutics can also alter blood volume, so in either case the miRNA measured can be skewed
Even with a standard set of steps that outlines how to isolate miRNAs from serum or sample, there are still a lot of knowledge that has not been sorted out. For instance, there is the challenge of choosing which miRNAs, out of the about 2000 miRNAs discovered, should be used as biomarker and for which diseases, given that there are some miRNAs, such as miR-21 which upregulation has been associated with colorectal, lung ,breast, prostate, liver, esophageal and endometrial cancers. There are also confounding factors that may influence how we establish its causal relationship with a disease, such factors can be age, gender, ethnicity, drugs or smoking, gut microbiome composition or diet; for example, what would be the real prognostic value of a certain or group of miRNAs for a childhood cancer instead of another age group’s cancer
Additionally, although miRNA promises a lot of diagnostic potential for the world’s most challenging diseases, this should not be confused with exploiting it as the sole predictor for pathogenesis. This is because phenotype is influenced by genotype and environmental factors, so all three variables should be taken into account in order to perform a disease diagnosis. With the intervention of artificial intelligence in medicine, next generation/deep sequencing should be taken as yet another tool available for the medical personnel to consult that’d aid in prognosticating a patient’s disease. This novel tool should be coupled with other definite testst and/or electronic health records that should be comprehensive more than ever, including information such as precise family history and genetic risk factors, environmental factors, or other X-RAY scans which information unable to be seen by the naked eye can now be seen by convolutional neural networks, among other diagnostic methods. Not only that, miRNAs are not the only circulating RNAs that should be the focus of researchers, as there yet a lot of non-coding RNAs yet to be discovered which could become even better biomarkers
For medicine, countless opportunities are being opened for the future of medicine
Therapeutic means for miRNA regulation
For a disease to be thoroughly defeated, its early diagnosis should be coupled with the pertinent treatment, which opens to yet another world of possibilities available where miRNAs can once again take the spotlight. This is because, as mentioned previously, these tiny ncRNAs have been shown to be involved in the regulation of mRNA expression as well as they’re able to reprogram molecular pathways in diseases like cancer
miRNA therapeutics offer optimism for the future of medicine, hence it’s a field also worth diving deep into, given that a therapy based on the delivery of RNA-carrying vesicles might just offer a simpler and safer alternative to, for instance, stem-cell therapy; such advantage extends itself to the treatment of even neurological disorders as long as researchers find ways to deliver such RNA-carrying vesicles across the blood-brain barrier
Also, current therapeutic targets are mostly proteins that belong to either one of the following three protein classes: enzymes, receptors and transporters. This has put a burdensome challenge to research and drug discovery given the large amount of resources needed to be invested (money in the scale of billions of dollars and longitudinal studies that may span decades), coupled with the natural challenges that drugs’ R&D carry with themselves (the need to follow Lipinski’s Rule of Five11 as well as making the drug actually modulate the target protein) and the very strict regulatory mechanisms that ensure drug safety; all in all, the end result is that the number of protein drug targets is limited; although the human genome encodes 100000–200000 proteins, it has been shown that only 207 proteins are targeted by FDA-approved small-molecule drugs. Furthermore, it is estimated that only 600 disease-modifying protein drug targets exist
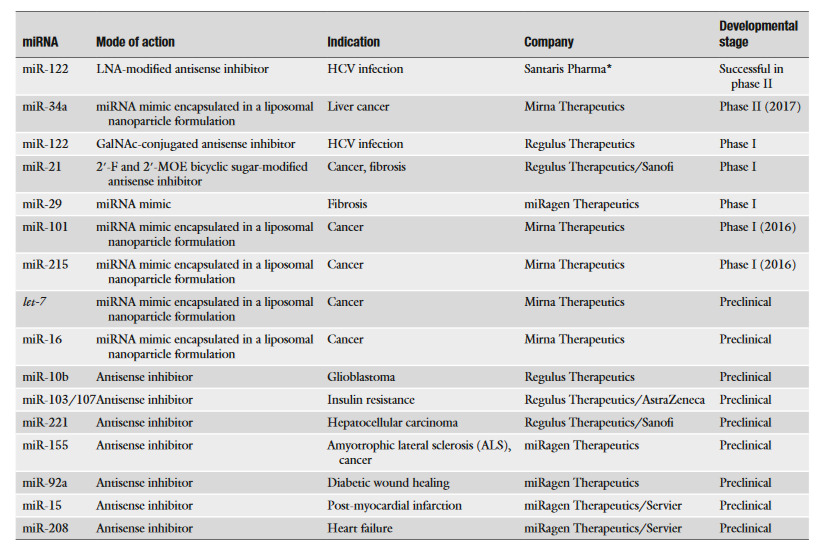
Another advantage is that given how miRNA biomarkers can be employed to develop a particular miRNA expression profile for a particular disease, this can also open the possibility of a personalized therapy based on such miRNA signature through either delivery of antisense miRNA strands to inhibit specific upregulated miRNAs or miRNA mimics to balance out specific downregulated miRNAs; [Table 1]summarizes some companies that are currently developing such strategies to diseases like cancer, HCV infection or heart failure. miRNA therapeutics hence may contribute to the foundations of personalized medicine by allowing targeted therapies, which are needed in disease like cancer so that specific oncogenic pathways are interfered, thus improving patient survival and care.
Finally, miRNA deregulation may not only affect pathogenesis, but also influence in drug resistance. A number of published reports show that manipulating expression of specific miRNAs can alter the drug sensitivity or that miRNAs are themselves biologically involved in the body’s resistance response, and this kind of phenomenon doesn’t restrict itself to cancer pathologies, but is also associated with drug resistance in treatment of conditions such as epilepsy, multidrug-resistant tuberculosis, and insulin sensitivity
Given such premise that shows yet another potential of miRNAs, now in the field of therapeutics, it is worth exploring how miRNAs can be delivered to the patient. There are several strategies that target putative miRNAs to treat a disease. Among them, there are the following:
Delivery of miRNAs mimics or antisense oligonucleotides
A therapy based on the delivery of nucleotides is an active area of drug development designed to treat a variety of gene-specific diseases
The use of oligonucleotides is inspired by the fact that while a deregulation of miRNAs has been causally correlated with a disease’s development, a compensation for such deregulation (for instance, inhibiting upregulated miRNA or enhancing downregulated miRNA) has also been shown, mostly in animal models or cellular assays, to alleviate the phenotype expressed by certain disease. For instance, in an infectious disease like tuberculosis,
Of course, it’s worth mentioning that these research results have been limited into either experimenting with animal models or cell lines, so they don’t necessarily translate into human models; nonetheless, an overall idea is that there are clear prospects to explore furthermore in the field miRNA therapeutics and strategies to compensate for a miRNA deregulation to alleviate the symptoms of a disease.
Also, the delivery of oligonucleotides, such as with liposomal nanoparticles, is a field that requires further research and development in order to overcome pharmacokinetic challenges and issues like poor permeability to a cell’s insides, where the target miRNA is located
Disruption or enhancement of a miRNA’s pathway of biogenesis
There are several strategies designed to modulate target miRNA’s concentrations, and before going through these strategies it’s worth first getting an idea of the causes of miRNA dysregulation (please examine
For instance, in miRNA biogenesis, as shown in Figure 7, molecules can intervene during DICER-mediated pre-miRNA processing in order to either upregulate a miRNA through enhancing its maturation, otherwise inhibit its maturation, thus reducing its expression levels. Targeting pre-miRNA comes with its advantage due to its relative bigger structure than miRNA, which increases its ligandability, thus increasing the change of finding a molecule to attach to them
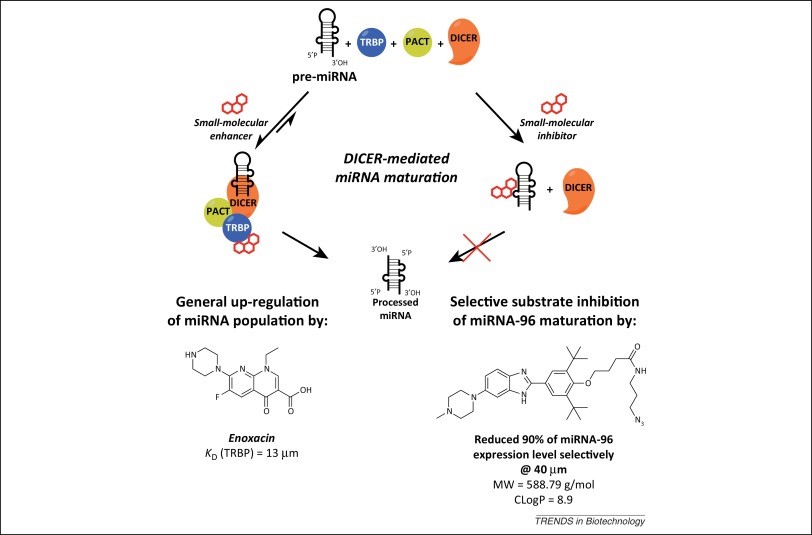
An alternative to the above is targeting the miRNA-AGO2 complex through anti-miR-AGOs. In theory, these should have better pharmacokinetic properties than oligonucleotides given that they don’t target miRNAs per se, but rather AGO2 protein’s active site with the miRNA of interest identified through a specific oligonucleotide sequence, which altogether leads to the development of more hydrophobic molecules with lower molecular weight than the mentioned oligonucleotide inhibitors
Epigenetics also offer an alternative for miRNA therapeutics. This is considering the reported evidence of how microRNA expression can be affected by changes in the epigenetic program. In theory, the existence of epigenetic drugs, such as DNA demethylating agents and histone deacetylase inhibitors, which are able to reverse an aberrant methylation or acetylation status, raises indeed the intriguing possibility to regulate microRNA levels, for example to restore the expression of tumor suppressor microRNAs, thus reverting a tumoral phenotype
miRNA editing: miRNA and CRISPR
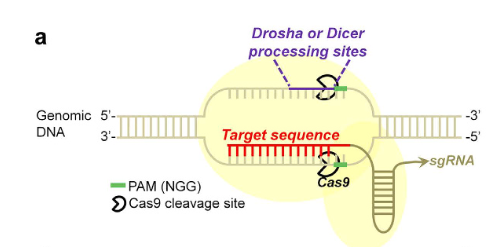
DNA may also be a therapeutic target regulating miRNAs. Given the high prospects displayed by the CRISPR – CAS13a system in nucleic acid detection, the CRISPR-CAS system wouldn’t be exempt from also being a therapeutic platform with great potential to be harnessed, given that it may compensate for the flaws of an oligonucleotides-based approach.
For example,
Of course, one such limitation that becomes apparent is that CRISPR-CAS9 seems to be focused only on knocking down a gene loci that leads to the downregulation of a desired miRNA, however, there needs to be further development in the area of how a CRISPR-Cas system can also upregulate miRNAs, as research has shown that upregulation of selected miRNAs can also reverse the phenotype of a disease, as shared previously (please revisit Table 3).
All in all, it could be the case that with enough research and development, the CRISPR/Cas9 system, or maybe another system that employs a novel nuclease yet to be discovered, could be a more efficient, specific, economic, convenient, and stable technology for knocking down miRNA, so that such therapeutic platform can be accessible for the general population instead of very technical settings like laboratories, limiting miRNA therapeutics to researchers only.
Dietary miRNA
“Let food be thy medicine and medicine be thy food” said Hippocrates. A disease prognosis is done better if coupled with a lifestyle that takes preventive measures towards a disease. Because it’s more than well shown that miRNAs regulate gene expression, their presence in exosomes in food like milk, ginger, mushrooms, grapes, etc., have attracted researchers’ attention to see how such RNA-carrying extracellular vesicles can affect the host, particularly the gut microbiome10. If research in this field is ripened, and by ripened we mean a thorough understanding of the gut microbiome and how miRNA carrying-exosomes interact with each other, theoretically, it’s possible to bioengineer exosome-like nanoparticles derived from plants or the food mentioned above for them to carry RNA (and of course maybe proteins and lipids) for therapeutic purposes for certain disease
Challenges of miRNA therapeutics
Just as how challenges arose from miRNA diagnostics, miRNA therapeutics also shows a lot of barriers to surpass. Despite the many opportunities that may burst forth from the ongoing research about ncRNAs, particularly miRNAs, there are diverse challenges, some of which have been shared sporadically in the sections above, such as the design of ASOs high ligandability and vesicles of high permeability.
Also, perhaps an advantageous, yet also an inconvenient feature of miRNAs is their ability to simultaneously target multiple genes, which has made them an attractive alternative to the ‘one target, one drug’ model, but it also decreases their overall disease specificity and increases the likelihood of potential off target effects
Another such double-edged-sword quality is miRNA’s great tissue/cell specificity, which raises a challenge for drug makers because the therapy must be targeted not only towards a specific miRNA, but a specific miRNA of a particular cell type, as noted by researcher Andrew Baker, a molecular biologist at the University of Edinburgh. This was regarding a miRNA he researched, miR-21, which was associated with plaque build-up in arteries and have been found seven times more abundant in peripheral arteries clogged with atherosclerosis than in healthy arteries. He noted that miR-21 is expressed in virtually every cell. If there is a broad, unfocused attempt to suppress such omnipresent molecule, then once again off target effects with unintended health consequences might occur
Also, as with miRNA being as biomarkers, it is always worth noting miRNA therapeutics is not complete if miRNA is the only point of focus. Once again, phenotype is the result of genotype and environmental factors, so for a disease therapy to be thorough, changes in the environment, lifestyle, diet, among other factors, must be taken into account.
As for drug targets, it’s also worth saying that miRNAs shouldn’t be treated as the only molecule to invest the future on. Protein, lipids, other circulating, noncoding-RNAs or molecules yet to be discovered all can become better therapeutic targets or biomarkers to treat diseases that nowadays ravages humanity, whether it is a cancer or pathogen-driven.
However, all in all, miRNAs seem to be a possible candidate to becoming the next blockbuster in medicine, with high prospects of revolutionizing (maybe together with the CRISPR-CAS system) disease prognosis and therapy.
Afterthoughts
Despite sharing some findings regarding miRNAs and their role in disease prognosis and therapy, may main goal is to inspire the reader to choose a path of R&D for his or her future. I also want to share a view regarding how artificial intelligence can play a pivotal role for bringing such technology to each household in order to democratize medicine and allow the general population to become healthier.
Such view is based on a product small and easy to use so that the average household can have access to it and employ it for a constant monitoring of the family members’ miRNA expression patterns.
- It may resemble an apparatus similar to miRoculus (or maybe miROculus themselves) or maybe MinION Figure 9 12 designed by Oxford Nanopore Technologies, which is a very small device used to perform DNA or RNA sequencing.
-
Whether the device is like miROculus or MinION, the inner architecture can be adapted from either the CRISPR-LbuCas13a detection platform by
or the CRISPR/Cas13a-Powered electrochemical microfluidic biosensor by , so that a miRNA diagnostics can be performed from a simple blood sample loaded to this small device. This is in order to reduce the cost of reagents as wells as facilitate the procedure of diagnosis. -
The device is then connected to a terminal, maybe a PC or personal laptop, to upload data about the identified miRNA expression profile. This data could also be shared through the cloud with a medical team or professional for a more thorough analysis. Either way, such data, coming from a blood sample that underwent CRISPR-CAS diagnostics for identifying a miRNA expression pattern, will act as input towards a deep neural network that’s able to prognose a disease from such. For a depiction of a possible network that performs such miRNA-disease associations, please examine
, where they share an implementation of a MiRNA-disease Association Prediction (MAP) network that’s constructed by combining miRNA-gene associations, protein-protein interactions (PPI), and gene-disease associations collected from databases to form a heterogeneous tripartite network that’s then reduced to a miRNA-disease bipartite network through a network diffusion algorithm that performs a random walk starting at the known disease genes to rank miRNA-disease associations (Figure 7). The output, hence, can resemble Figure 11, given that’ll be user friendly and easier to understand.
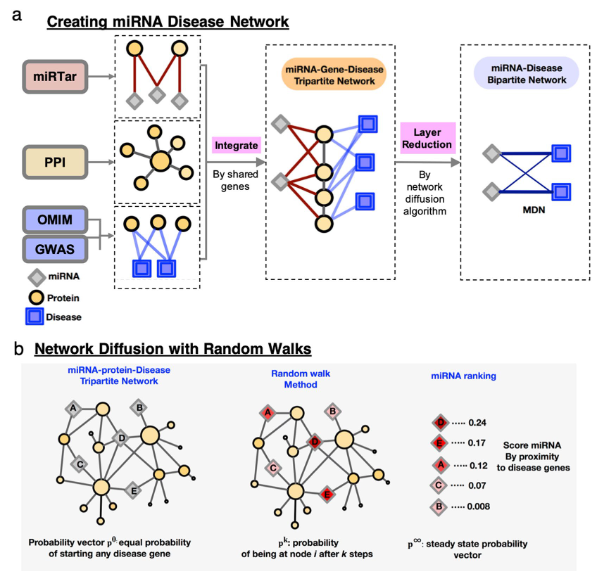

I envision that an addition to such network would be the prediction of also putative miRNA targets that may aid the relevant medical personnel to design the most appropriate therapy (which may also be based on a CRISPR-Cas system). Also, the network can be further trained to prognose a disease from other covariates such as age, gender, ethnicity, gut micriobiome composition, among others to perform a more precise diagnosis.
All in all, as said before, miRNAs can be next blockbuster in medicine that may aid in revolutionizing the field of diagnostics and therapy.
-
For further inquiry into these networks, the following website for learning can be visited: https://distill.pub/ ↩
-
The cited source is the original document in Chinese published by Prof. Zhu. For an English version in the format of an e-book, please visit: https://dm.ai/ebook/ ↩
-
Along with Yoshua Bengio and Yann LeCun. ↩
-
For further exploration on such topic, I recommend reading Deep Medicine by Dr. Eric Topol. ↩
-
Please see The Human Genome Project: https://www.genome.gov/human-genome-project ↩
-
It’s quiet controversial to suggest that all cells, with few exceptions, carry the same DNA sequence. From (Eric, 2015) in The Patient Will See You Know, we have to take into account that different cells from the same individual might carry mutations in their gene sequences. There is another slight issue which is that there are still genes yet to be identified. But these are details that can be skipped if we want to have a basic understanding of miRNA’s relevance. ↩
-
Alternatively, these stem cells are the ones employed for genetic engineering thanks to them being partially differentiated or completely undifferentiated, which allows them to be used for experimenting as mimics of human organs, called organoids (MIT Technology Review, 2019), for instance. ↩
-
To gain a deeper understating from such brief explanation, please visit https://phet.colorado.edu/en/simulation/gene-expression-essentials for an interactive simulation, as well as read further on the literature about gene expression. ↩
-
Cas9 differs from Cas13. The former nuclease is guided by an sgRNA towards a target sequence in the DNA; subsequently, Cas9 makes a double strand break in the DNA that’s followed by endogenous repair mechanisms that may result in gene knockout via frameshift mutation or gene knock in if a DNA template is present (Synthego, 2019). Though the phase of recognition of a target sequence is similar, the latter nuclease’s cleavage mechanism is exploited to indicate the presence of a desired nucleic acid. ↩ ↩2
-
There has been recent attention focused on the gut microbiome; for a deeper insight please visit: https://www.nature.com/collections/eccfeecfae ↩ ↩2
-
A set of rules that’d enable a drug to be assimilated by the body in order to target a protein. Such drug should have the following qualities: no more than 5 H-bond donors, 10 H-bond acceptors, the molecular weight is no greater than 500 and the calculatedLog P (CLogP) is no greater than 5 (or MlogP.4.15) (Christopher, Franco, Beryl W., & Paul J., (2001) ). ↩
-
Please visit: https://nanoporetech.com/products/minion ↩